|
|
|
Medical Pharmacology Chapter 36: Antiviral Drugs
Antiviral Drugs
More about Influenza Viruses.
Orthomyxoviruses and paramyxoviruses are the two families originally described by "myxoviruses".4
The origination of the myxoviruses name was based on the finding that virions associated with influenza and some other enveloped, negative-strand RNA viruses bind to sialic acid residues in mucoproteins. (Here "ortho = correct and para = alternative).
The Orthomyxoviridae family not only includes influenza viruses A, and but also two tick-borne mammalian viruses in the Thogotovirus genus (from the Kenyan Thogoto forest where the virus was first isolated) and a virus (Isarvirus genus) which infects Atlantic salmon.
 |
|
![]() Influenza
type A viruses cause most flu epidemics and are commonly found in both
avian and mammalian species.4
Influenza
type A viruses cause most flu epidemics and are commonly found in both
avian and mammalian species.4
By contrast, influenza type B is found only in humans; whereas, type C infects both humans and pigs.
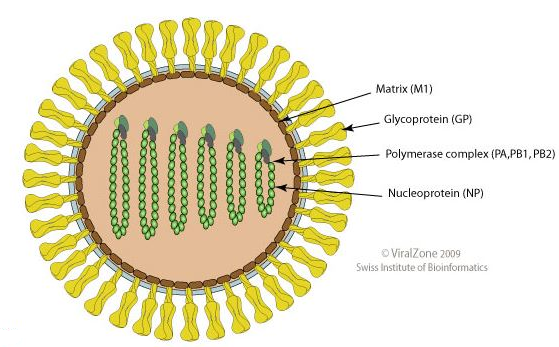
Orthomyxoviruses exhibit both common structure and replication process.4
Their negative-strand RNA is comprised of 6-8 gene segments, individually associated with helical nucleocapsids.
These nucleocapsids are packaged in a host cell plasma membrane-derived lipid envelope forming the virion.4
|
|
|
Influenza viral genome segments:
![]() 11
viral proteins are produced by influenza type A viruses.4
11
viral proteins are produced by influenza type A viruses.4
9 of these 11 are packaged in virions.4
Genome segments 1 though 6 each code for one protein as follows:
3 RNA polymerase subunits (PB2, PB1, and PA)
Gycoprotein hemagglutinin (HA) envelope glycoprotein
Nucleocapsid protein NP and an additional envelope glycoprotein, neuraminidase (NA).4
For some influenza A isolates, an additional small protein may also be encoded, PB1-F2.
mRNAs transcribed from genome 7 and 8 may be spliced which results in 2 distinct mRNAs, coding for different viral proteins.4
Genome segment 7 encodes the sequence for matrix proteins (M1) as well as a third envelope protein, M2.
Genome segment 8 encodes for NS1 and for NS2, both nonstructural proteins.
All of these proteins have important roles in viral replication.4
|
|
|
Role of hemagglutinin in influenza viral replication:
Influenza virus has an ability to agglutinate red blood cells.4
This action appears mediated by influenza hemagglutinin (so named) protein (HA), a protein which binds to sialic acid-containing receptors.
These receptors are not only localized on red blood cells but also on most other cells.
HA forms a "trimer" in the virus enzyme and is key for virion binding to the cell surface and also for viral genomic entry into the cell.
![]() Entry
of the virus into the cell occurs by a fusion pathway.4
Entry
of the virus into the cell occurs by a fusion pathway.4
HA, a type I transmembrane protein, after synthesis, is first localized in endoplasmic reticulum membrane and later transported by the Golgi apparatus to the plasma membrane where it can be incorporated into virions.4
The role of influenza hemagglutinin protein, HA, in membrane fusion is dependent on cellular proteinases that mediate cleavage of HA into two subunits.
This cleavage is necessary for membrane fusion activity.
The first subunits, HA1, is the surface subunit which contains the sialic acid binding region.
HA2 is localized in the virus envelope and contains the fusion peptide.
Initially, the fusion peptide resides in an inaccessible hydrophobic region of HA, close to the viral envelope lipid membrane.4
The fusion activation process is initiated subsequent to virion binding to cell receptors.
In this step, virions into the cell by means of an endosomal vesicle and with a decrease in endosomal pH (increased acidity), a change in HA confirmation is induced.
One consequence of this change in conformation is the transition of the fusion peptide out of the inaccessible, hydrophobic region towards the endosomal membrane.
There, this new HA confirmation allows insertion of the protein into the membrane.
Concurrent with that step is a change in location of HA2, anchored in the viral envelope, and now moved closer to the extended N-terminal region.
|
|
|
In this stage, one end of HA1 is associated with the endosomal membrane with the other end associated with viral envelope.
The described conformational change allows the approximation of these two membranes and membrane fusion.4
|
|
|
|
|
|
|
|
|
|
|
|
|
|
This Web-based pharmacology and disease-based integrated teaching site is based on reference materials, that are believed reliable and consistent with standards accepted at the time of development. Possibility of human error and on-going research and development in medical sciences do not allow assurance that the information contained herein is in every respect accurate or complete. Users should confirm the information contained herein with other sources. This site should only be considered as a teaching aid for undergraduate and graduate biomedical education and is intended only as a teaching site. Information contained here should not be used for patient management and should not be used as a substitute for consultation with practicing medical professionals. Users of this website should check the product information sheet included in the package of any drug they plan to administer to be certain that the information contained in this site is accurate and that changes have not been made in the recommended dose or in the contraindications for administration. Advertisements that appear on this site are not reviewed for content accuracy and it is the responsibility of users of this website to make individual assessments concerning this information. Medical or other information thus obtained should not be used as a substitute for consultation with practicing medical or scientific or other professionals. |